 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free! Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
> Insights > A year later: What we know about SARS-CoV-2 infectivity and growing concerns of emerging variants It has been a year since the COVID-19 pandemic began and plethora of research into SARS-CoV-2 has led to development of vaccines and therapeutics. In this article, we review what we’ve known about SARS-CoV-2 and COVID-19, the developments in fighting the pandemic and the threat of emerging variants.
The article is organized into four parts: • Part 1: COVID-19 and SARS-CoV-2 • Part 2: S protein: key to the pandemic • Part 3: Understanding Infectivity • Part 4: A new frontier of emerging variants |  |
The answers to the following questions will be cleared after reading the text:
•What is the mechanism of fusion protein?
•Why S protein is the key to the pandemic?
•What does the Super stable S protein trimer mean?
•How SARS-CoV-2 infects the host?
•Are there any other host receptors besides ACE2 protein?
•What are the major SARS-CoV-2 variants in circulation worldwide?
•Could the emerging mutations impact the transmission, diagnosis, therapeutics and vaccination of SARS-CoV-2?
You can also find the answers in the video:

COVID-19 to date has infected 100 million people and led to 2.2 million death worldwide (Figure 1A). The viral agent of this pandemic SARS-CoV-2 is an enveloped virus with a single strand positive sense RNA genome.
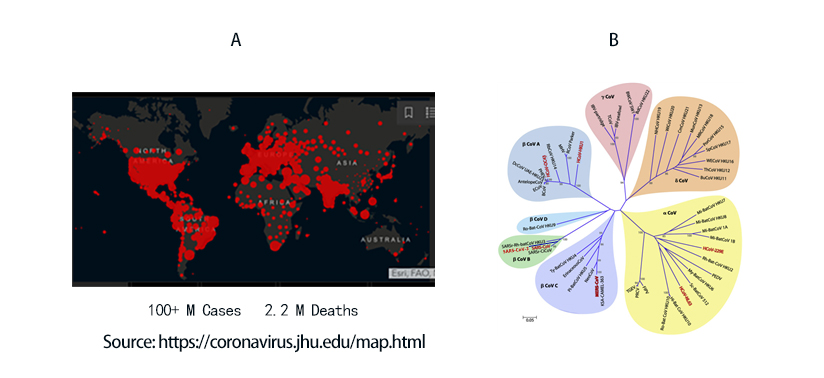
Figure 1. A) Global map of SARS-CoV-2 infection to date. B) Phylogenetic tree of coronaviruses. SARS-CoV-2 belongs to -coronavirus clade in the B lineage.
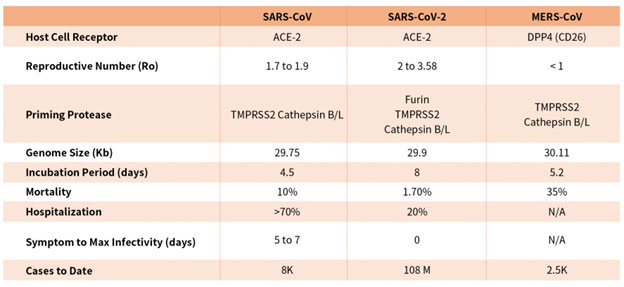
Coronaviruses are a family of viruses with distinct morphology of crown like projections infecting birds and mammals and are divided into four genera: a, b, g and d(Figure 1B). Human coronaviruses were known to typically result in mild upper respiratory tract (UTR) infections until the outbreak of the SARS epidemics of 2002. The SARS followed by MERS epidemic showed these coronaviruses ability to jump hosts and cause serious lower respiratory tract infection. While SARS-CoV, SARS-CoV-2 and MERS-CoV belong to the b-coronavirus family, key differences in SARS-CoV-2 structural proteins has enabled it to rapidly explode in transmission and gripped the world with a once-in-a century pandemic (Table 1). Rapid research into the structural basis of SARS-CoV-2/ host cell interaction and mechanism of viral entry has only been possible based on previous SARS and MERS outbreak studies. Although both SARS-CoV and SARS-CoV-2 bind the same receptor on host cells via similar mechanisms, there are considerable differences accounting for high transmission rates of SARS-CoV-21. Coupled with longer incubation and lower hospitalization rates, a novel furin cleavage site as well as no delay between symptom onset and maximum infectivity (Table 1), SARS-CoV-2 has been able to spread rapidly around the globe compared with prior coronavirus pandemics.
Table 1. Characteristics of the three recent coronavirus outbreaks2, 3
However, rapid advancement in understanding the structural underpinning of SARS-CoV-2 infectivity and transmissibility has yielded promising therapeutics and vaccine candidates. Continual examination and study of viral mechanism and impact of emerging mutations are key to turning the tide on this pandemic.
The SARS-CoV-2 viral particle consists of a lipid of lipid bilayer containing membrane (M) protein, envelop (E) protein, nucleocapsid (N) protein and the Spike (S) protein as shown in Figure 2.
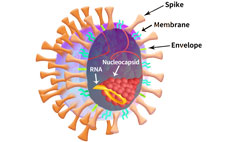
Figure 2. SARS-CoV-2 particle schematic.
Among all of the structural proteins, viral fusion protein plays a critical role in facilitating envelop viral particle entry and by proxy virus genetic material into host cell4. In the case of SARS-CoV-2, the S protein is the viral fusion glycoprotein trimer which binds to host cell receptor and enables viral entry. After entry, host cell machinery is hijacked for replication, transcription and assembly of viral particle propagating its spread. Furthermore, viral fusion protein when expressed in host cell membrane can induce cell-cell fusion contributing to viral spread and virulence4. In particular, the thermodynamic barrier of merging enveloped virus membrane to host cell plasma membrane is overcome by the kinetic energy stored in the fusion protein4. The prefusion state of the fusion protein in most cases is metastable and triggering events convert it to pre-hairpin intermediate which exposes the hydrophobic fusion peptide embedding into host cell membrane4. The fusion proteins can be divided into 3 classes based on their structures and triggering mechanism. Class I is primarily comprised of helices, while class II consists primarily of b sheets and class iii contains a mixture of a helices and b sheets4.
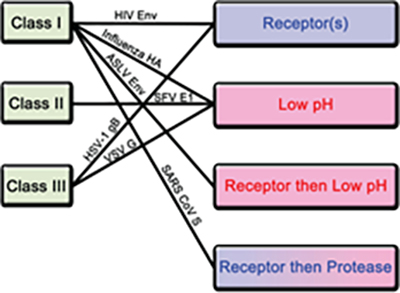
Figure 3. Classification of fusion proteins and triggering mechanisms with examples of corresponding viruses.
There are 4 known triggering mechanism for fusion protein including receptor binding, low pH, receptor binding followed by low pH and receptor binding followed by protease cleavage (Figure 3). Aside from class II, both class I and III fusion proteins are triggered by multiple mechanisms. The S protein of novel SARS-CoV-2 is a class I fusion protein triggered by receptor binding and protease cleavage5.
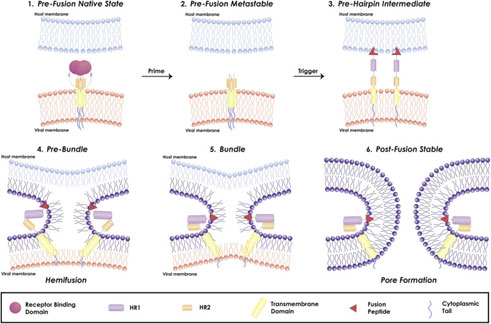
Figure 4. Mechanism of viral fusion entry primarily for class I fusion proteins. Step 4-6 applies to other classes of fusion proteins as well4.
The mechanism of a class I fusion protein begins with the pre-fusion state where the receptor binding subunit is clamped to the fusion subunit. The fusion protein can bind to a single or multiple receptor proteins. Triggering of the fusion protein after receptor binding and protease cleaving, enables the receptor-binding subunit to move out of the way allowing the fusion subunit to form a pre-hairpin with target membrane via fusion peptide4 (Figure 4). Subsequently, the pre-hairpin folds back causing N and C alpha helix heptad repeats to form a 6-helix bundling pulling the two membranes through close apposition, hemi fusion and fusion pore formation as shown4 (Figure 4). Although enveloped viruses utilize variety of different types of fusion proteins and triggering mechanism, all converge in these three last steps to gain entry into the cells. Examination of the viral fusion entry for SARS-CoV-2 is critical to developing effective therapies against COVID-19 and designing effective vaccines.
Comprehensive reagents are available at ACROBiosystems
ACROBiosystems has developed a comprehensive line of critical reagents, including recombinant proteins, antibodies, kits, beads and so on, covering the critical targets of SARS-CoV-2. We hope that we can help to accelerate the research and development of SARS-CoV-2 related therapeutics, diagnostics, and vaccines with these high-quality products and advanced technical support.

Prior to SARS-CoV-2 outbreak, the S protein of SARS-CoV had been studied extensively. The S protein among other structural proteins of SARS-CoV-2 was found to be highly immunogenic and effective neutralizing antibodies recovered from SARS patients were directed at S protein6, 7. Therefore, research into vaccine and therapeutics against SARS-CoV-2 were immediately focused on the S protein. Although the four structural proteins of the novel SARS-CoV share more than 90% in sequence identity with SARS-CoV-2, S protein consisting of S1 and S2 subunits diverged with only 77% sequence similarity1.

Figure 5. Structural schematic of S protein between SARS-CoV and SARS-CoV-21.
The S1 subunit contains the NTD or the N-terminal domain and Receptor binding domain (RBD) which binds to the human angiotensin converting enzyme-2 (ACE2) primarily through direct interaction with the receptor binding motif (RBM). The S2 subunit comprises of the fusion peptide, heptad repeats, transmembrane region as well cytoplasmic domain (Figure 5). The high transmissibility of SARS-CoV-2 compared with SARS-CoV has been posited to occur due to the insertion of a polybasic cleavage site between S1 and S2 subunits which reduces stability of S protein and facilitate conformational change required to bind the ACE2 receptor1. In fact, lentivirus presenting S protein in a pseudoviral entry assay shows that protease cleavage of S protein occurs during viral entry8. Mutating the putative polybasic cleavage site on S protein abrogated the cleavage (Figure 6A).
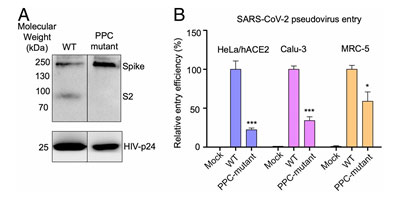
Figure 6. S protein cleavage occurs and necessary for viral infection. A) Mutation of furin (PPC) site abolishes cleavage. B) PPC mutant have lower viral entry efficiency compared to WT.
Furthermore, viral entry efficiency in ACE2 expressing HeLa cells and lung cancer cell lines was significantly lowered in cleavage site mutant of S protein8 as shown in Figure 6B. These results indicate the necessity of cleavage of S protein for viral entry.
Deeper understanding of the structural underpinnings of S protein function in viral attachment and entry has been accelerated by cryo-EM structures of S protein. Structural analysis of the S protein revealed a trimeric structure with a central stalk consisting of 3 S2 subunit as shown (Figure 7A) which is topped by 3 S1 subunits9. The S1 subunit is dynamic with conformation changes oscillating between open and closed states. In the S1 open conformation the RBD is accessible to binding via ACE2 receptor. These movements and rearrangements by RBD and NTD domains of S1 subunit are driven by energetics of ACE2 binding9. ACE2 binding alters the open RBD conformation by a rigid -body rotation of the domain moving it further 5.5 Å away from trimer axis while the NTD domain moves 1.5 to 3 Å9. The RBD associated subdomain 1 moves about 8 Å while the NTD-associated subdomain 2 moves by 3 Angstrom (Figure 7B).
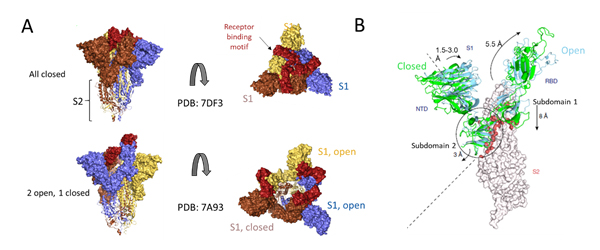
Figure 7. Cryo-EM structure of S trimer. A) Depiction of S1 subunits in open and closed RBD conformation as surface representation and S2 subunits in ribbon form. B) Changes in S1 subunit post ACE2 receptor binding.
The NTD associated subdomain 2 interacts with a short helix and loop of S2 subunit via series of stacking in the closed conformation (Figure 7B). These interactions are lost in the open conformation leaving a channel in S2 that is disordered and exposed for the next step of membrane fusion9. The major rearrangement of the S trimer is fueled by the energetics of ACE2 binding events. ACE2 binding favors RBD open conformation and sequential opening and binding of RBD in the S trimer leads to a fully open state leading to substantial reduction in contact between S1 neighbors and S2 trimeric core. The fully open state leaves a 50 Å diameter around the trimer axis that is 65 Å deep9. Furthermore, this new open structure leads to solvent exposure of the fusion peptide on S2 as well as the major helical rearrangement leading to host cell fusion.
Due to the dynamic nature of S protein, domains in the S1 subunit and S2 subunit comprises of centroids with various angles and dihedrals10 (Figure 8A). This means mutations at sites intersecting these centroids will affect the conformation landscape of the S protein impacting vaccine and therapeutic development. Additionally, prefusion stable S protein are needed to increase recombinant protein expression yield with homogenous preparations11, 12. Proline substitutions (2P and 6P) have been identified to stabilize loops and cap helices in the prefusion state (Figure 8B) as well as introduction of disulfide bonds prevents conformational changes11.
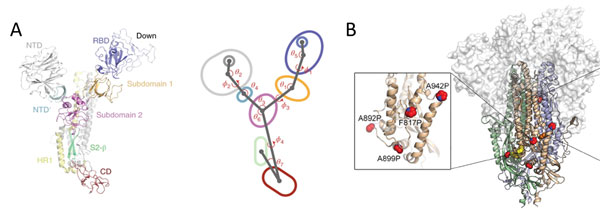
Figure 8. A) S protein is highly dynamic structure with multiple centroid rotating along various angles and dihedrals10. B) 6 Proline mutations in S protein timer stabilizes the prefusion state.
Featured super stable S protein trimer from ACROBiosystems
ACROBiosystems provides a super stable S protein trimer for vaccine and therapeutic development. The trimer contains a T4 fibritin trimerization motif in addition to the 6 prolines substitution to stabilize the trimeric prefusion state. Additionally, furin cleavage site has been mutated via two alanine substitutions. We have isolated high purity S trimer with high homogeneity. The S trimer particle structure was verified via negative stain electron microscopy and matches reports from the literature. Additionally, performance testing of our Super stable S trimer with WT S trimer and 2 proline substituted S trimer along with S trimer from a competitor showed ACROBiosystems Super stable S trimer with 6 proline mutation to consistently have the highest signal to noise ratio. Such high performance will greatly improve development of diagnostic kits as well as vaccine immunogenicity evaluations.
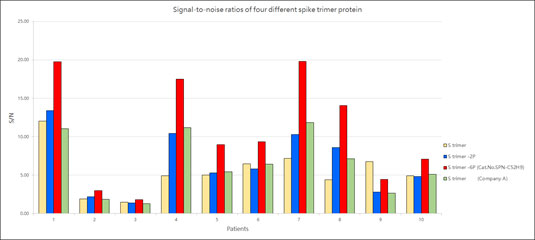
Figure 9. Signal-to-noise ratios of four different spike trimer protein
(Related article: Recombinant Spike Protein Trimer: more powerful than you think)
SARS-CoV-2, like its predecessor, primarily targets ACE2 receptor for viral entry and infection. Biologically, ACE2 functions to counteract the renin-angiotensin system to convert Angiotensin peptides which are hormones regulating vasoconstriction and vasodilation in cells. The ACE2 receptor is found in high levels in heart, kidneys and testis. ACE2 structure consists of the functional extracellular peptidase domain, followed by collectrin like domain or the neck domain, the transmembrane domain and 40 residue intracellular segment not resolved in these structures (Figure 10A). A protease cleavage site in the neck domain ACE2 enables secretion of active ACE2 peptidase. ACE2 dimerizes mediated primarily by the neck domain via H-bond and cation- interaction into an open and close conformation(Figure 10A)13. The open conformation of ACE2 binds to the RBD domain of S protein. Docking S protein trimer reveals that each ACE2 dimer can bind to two S protein trimers simultaneously as modeled (Figure 10B)13.
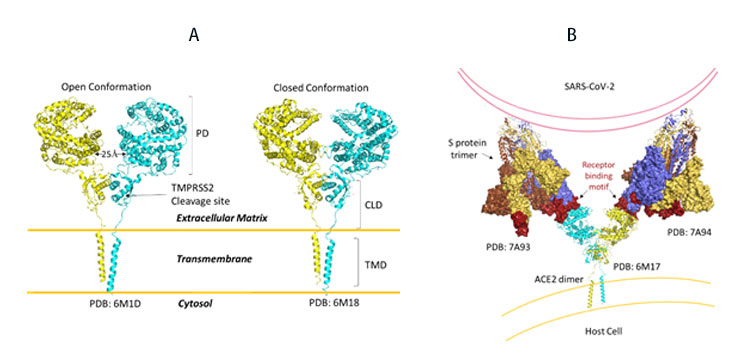
Figure 10. A) Full length structure of ACE2 in open and closed homodimer states. B) Modeled interaction of S trimer to ACE2 dimer during viral infection.
It remains to be seen whether clustering of S trimer and ACE2 dimers occurs and enable fusion of entry of SARS-CoV-2.
Upon closer examination of residues that are involved in RBD-ACE2 interaction, the receptor binding motif (RBM) forms physical contact with ACE2 receptors. Key residues on the RBM within the RBD make physical contacts with ACE2 receptor and lead to large buried surface of 1,687 Å2 formed by interactions of 17 RBD residues to 20 ACE2 residues (Figure 11A).
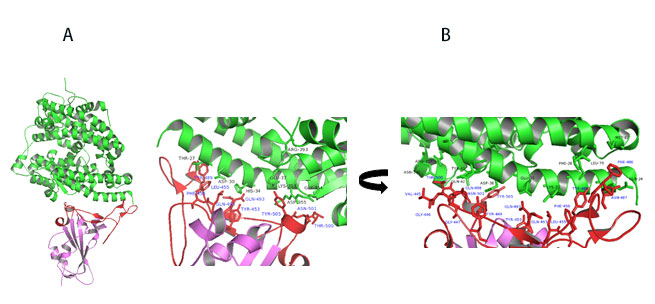
Figure 11. A) Co-crystal of RBD (purple) bound to ACE2(green). B) Polar interaction within the RBM that enable tight binding to ACE2. RBM is colored red. PDB: 6M0J
Key interactions are mediated by polar residues (13 H-bond and 2 salt bridges) similar to SARS-CoV1, 14. Particularly an extended loop region of RBD (containing b5, b6 strands and a4 and a5 helices and loops) forms a bridge with the a1 helix of ACE214 as shown in Figure 11A-B.
Receptors for Featured super stable S protein trimer from ACROBiosystems
ACROBiosystems offers ACE2 proteins with various species and tags to facilitate in-depth study of S-Protein/ACE2 binding mechanism and assessing efficacy of vaccines and therapeutics.
(Related article: Which animal model can help defeat COVID-19?)
In addition to ACE2, Neuropilin-1 receptor which binds to furin cleaved substrates has been shown to facilitate SARS-CoV-2 viral entry particularly in the nasal cavity cells. Expression of both Neuropilin-1 and ACE2 receptors in HEK-293T cells significantly improved viral entry than ACE2 alone(Figure 12)15. In fact, due to low level of expression of ACE2 receptor in respiratory and epithelial cells, Neuropilin-1 receptor is thought to promote entry of SARS-CoV-2 enabling cell, vascular and tissue penetration. ACROBiosystems offers his tagged and biotinylated Neuropillin-1 with high purity and bioactivity.
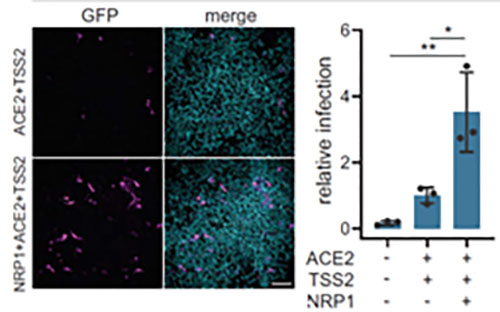
Figure 12. Neuropilin-1 and ACE2 co-expression dramatically improves viral entry
Recently S1 domain of SARS-CoV-2 has been showed to bind to Cholesterol and components of high-density lipoprotein (HDL)16 as shown in Figure 13A. Therefore, the HDL scavenger receptor B type 1 (SR-B1) has been implicated in enhanced ACE2 dependent viral entry as seen in the pseudoviral entry assay. Expression of SR-B1 improves viral entry while knockdown of SR-B1 worsens viral entry16 (Figure 13B-C). SR-B1 is co-expressed with ACE2 in pulmonary tissue and acts as a host factor promoting SARS-CoV-2 entry.
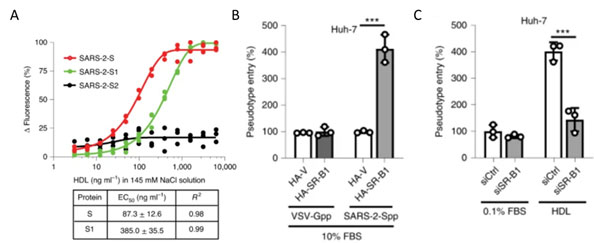
Figure 13. A) S protein S1 subunit binds to components of HDL. b) HDL receptor SR-B1 enhances SARS-CoV-2 entry.
Recently a large scale screening of 5054 human membrane proteins identified a panel of receptors that have strong affinity to SARS-CoV-2 spike protein and could point to alterative viral entry pathways that could explain disparate symptoms of COVID-1917. Other recent studies have reported other receptors, such as restriction enzymes and proteases, are also involved in SARS-CoV-2 infection18.
According to reference description, ACROBiosystems tried to verify binding affinity between SARS-CoV-2 S protein with the following human receptors:
Table. 2 Kd measurement of the interaction of SARS-CoV-2 S protein with human receptors
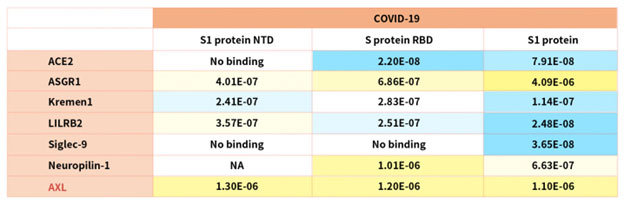
Notably, the latest research indicated that the tyrosine-protein kinase receptor UFO (AXL) specifically might interact with the SARS-CoV-2 Spike protein and take a key role in promoting viral infection of the human respiratory system.
(Related article: A summary of SARS-CoV-2 cell entry mechanisms)
ACROBiosystems is proud to partner with researchers at Beijing Institute of Biotechnology, Academy of Military Medical Sciences (AMMS) in uncovering the mechanism of this novel interaction which can be a novel therapeutic target. We are excited to continue partnering with leading SARS-COV-2 researchers in elucidation mechanism of viral infection and develop new therapeutics.

Mutations in the RBD have become a cause of concern as reports suggest higher transmissibility and reduced vaccine and antibody therapeutic efficacy in these strains.
As SARS-CoV-2 spreads globally, random mutations of viral genome are inevitable. Mutations in and outside RBD domain can significantly alter the conformation of S protein which needs to be monitored continuously for effect in transmissibility and severity of COVID-19. GISAID is tracking new mutation/clades of SARS-CoV-2 in the course of this pandemic.
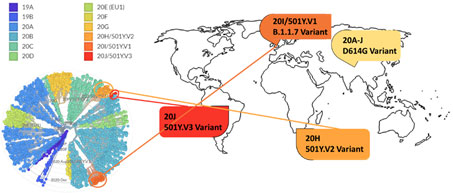
Figure 14. Phylogenetic clades of SARS-CoV-2 mutations with key variants around the world highlighted.
Next strain visualization of the SARS-CoV-2 mutation shows multiple clades of mutants emerging in 2019 and 2020 (Figure 14). However, mutations that lead to gain of functions are especially dangerous as it increases viral transmissibility and disease severity. To date the D614G variant is the dominant mutation found in virtually all SARS-CoV-2 infections and has been attributed to increase in SARS-CoV-2 transmission. Multiple explanation for high transmissibility of D614G variant has been shown including improvement in assembling of functional S protein on virion19 and equilibrium shift of RBD from 47% open to 95% open conformation in the mutant20. Recently three new variants have emerged: 501Y.V1/B.1.1.7 from UK, 501Y.V2/B.1.351 variant from S. Africa, and 501Y.V2/P.1 from Brazil. More recently, a US variant originating in west coast (B.1.429) has been identified and suspected in rapid increase of cases21.
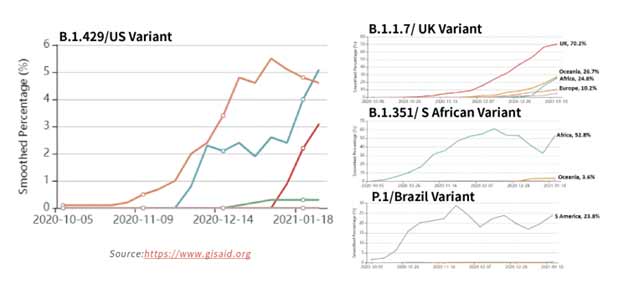
Figure 15. Tracking of variant percentages in SARS-CoV-2 samples as of Jan 28.
The spread of these strains worldwide (Figure 15) has been alarming due to suspected increase in transmissibility and potential resistance to vaccine induced/therapeutic neutralizing antibodies. The mutations details for the variants are shown in Table 3 below.
Table 3. Major SARS-CoV-2 variants in circulation worldwide21-23. Bold mutations are in the RBD and red colored mutations lie within the RBM.
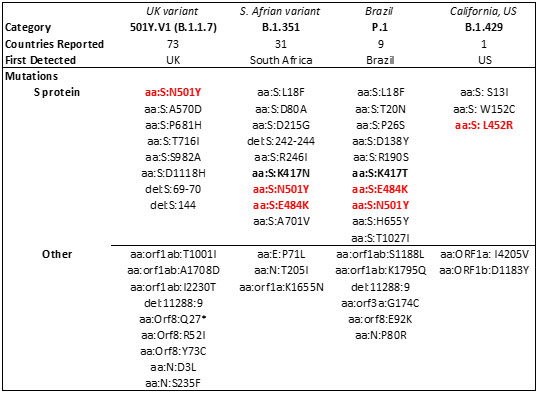
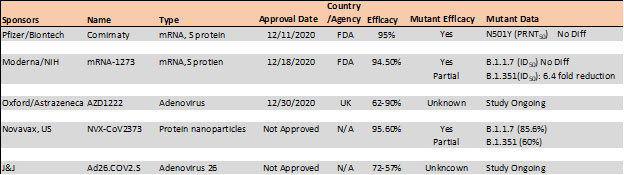
Mutations in the RBD have become a cause of concern as reports suggest higher transmissibility and reduced vaccine and antibody therapeutic efficacy in these strains. Recent report suggest, the B.1.1.7 variant is resistance to neutralization by antibodies targeting NTD of S protein and partially resistance to RBD mutants while showing modest resistant in convalescent plasma and vaccinated sera22. However, the B.1.351 has shown stronger resistance to both NTD and RBD antibodies and significant resistance (11-33 fold) in convalescent plasma ad vaccine sera (6.5-8.6). Results with Pfizer/BioNTech, Moderna and Novavax vaccines, the antibody titer shows no difference against B.1.1.7 variant while lowered efficacy has been observed against B.1.351 variant 24, 25.
Table 4. Vaccine efficacy analysis against emerging strains24-26.
The E484K mutation present in both S. Africa and Brazil variants is thought to contribute to resistance where neutralization reduction >10 fold was observed in some convalescent serum antibodies27. Another mutant, N501Y found in UK, S African and Brazil variant is thought to improve transmissibility of SARS-CoV-2 via strengthening ACE2 binding28.
In addition to effect in vaccination, these emerging variants have been found to lower potency of current antibody therapies against COVID-19. Two monoclonal antibody cocktail from Eli Lilly (bamlanivimab/estesevimab) and Regeneron (Casirivimab/imdevimab) have received emergency use authorization from FDA have been tested against these variants. In the case of Eli Lilly’s antibodies, bamlanivimab was susceptible to the E484K mutation in RBD leading to more than 100-fold reduction in neutralizing activity while estesevimab retained activity with only 4.4-fold decrease in neutralizing ability29. Regeneron’s casirivimab and bamlanivimab both showed little to on effect in potency against the B.1.1.7 variant while showing total or significantly abolished activity against B.1.347 variants.22
ACROBiosystems is going full steam ahead on developing a collection of recombinant antigens for these variants. The reagents can be used to evaluate the efficacy of the antibodies and vaccination.
Newly Launched product: Neutralizing antibody titer kit accelerating vaccines and diagnostics development
ACROBiosystems has developed multiple serological tests to track and fight the surging COVID-19 spread. Testing the levels of neutralizing antibodies against S protein in serum samples is critical in determining the efficacy of vaccines as well as a diagnostic measure of past infections. With the beginning of large-scale immunization with multiple vaccines and the rise of mutant variants, close monitoring of neutralizing antibodies in population and the impact of virus mutation is necessary for effective immunization campaign. ACROBiosystems offers neutralization kits as well as IgG, IgM titer detection kits. Our neutralization kit showed consistent detection of neutralizing antibodies against various dilution of COVID-19 convalescent serum. ACROBiosystems’ serology kits enable better understanding of neutralizing antibody mechanisms and help researchers develop strategies to improve vaccines and diagnostics particularly against emerging SARS-CoV-2 mutant strains.
reference
B, H.; H, G.; P, Z.; ZL, S., Characteristics of SARS-CoV-2 and COVID-19. Nature reviews. Microbiology 2020.
E, P.; M, K.; U, G.; DH, H.; N, P.; F, C.; M, S.; S, A. K.; L, S., Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. The Lancet. Infectious diseases 2020, 20 (9).
Z, Z.; X, L.; X, S.; W, W.; GA, M.; Y, Z., From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research 2020, 21 (1).
JM, W.; SE, D.; M, B.; K, S., Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Critical reviews in biochemistry and molecular biology 2008, 43 (3).
T, T.; M, B.; JA, J.; GR, W.; S, D., Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral research 2020, 178.
Buchholz, U.; Bukreyev, A.; Yang, L.; Lamirande, E.; Murphy, B.; Subbarao, K.; Collins, P., Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proceedings of the National Academy of Sciences of the United States of America 2004, 101 (26).
Traggia, i. E.; Becker, S.; Subbarao, K.; Kolesnikova, L.; Uematsu, Y.; Gismondo, M.; Murphy, B.; Rappuoli, R.; Lanzavecchia, A., An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nature medicine 2004, 10 (8).
J, S.; Y, W.; C, L.; G, Y.; Q, G.; A, A.; F, L., Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America 2020, 117 (21).
DJ, B.; AG, W.; P, X.; C, R.; SR, M.; PB, R.; JJ, S.; SJ, G., Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588 (7837).
R, H.; RJ, E.; K, M.; K, J.; V, S.; SMC, G.; M, K.; D, L.; R, P.; AL, H.; MJ, B.; BF, H.; P, A., Controlling the SARS-CoV-2 spike glycoprotein conformation. Nature structural & molecular biology 2020, 27 (10).
CL, H.; JA, G.; JM, S.; AM, D.; HC, K.; K, J.; KC, L.; D, W.; AG, L.; Y, L.; CW, C.; PO, B.; CK, H.; NV, J.; J, L.-M.; AW, N.; J, P.; N, W.; D, A.; JJ, L.; GC, I.; JA, M.; IJ, F.; JS, M., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science (New York, N.Y.) 2020, 369 (6510).
D, W.; N, W.; KS, C.; JA, G.; CL, H.; O, A.; BS, G.; JS, M., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020, 367 (6483).
R, Y.; Y, Z.; Y, L.; L, X.; Y, G.; Q, Z., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.) 2020, 367 (6485).
J, L.; J, G.; J, Y.; S, S.; H, Z.; S, F.; Q, Z.; X, S.; Q, W.; L, Z.; X, W., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581 (7807).
Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L. D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Meer, F. v. d.; Kallio, K.; Kaya, T.; Anastasina, M.; Smura, T.; Levanov, L.; Szirovicza, L.; Tobi, A.; Kallio-Kokko, H.; Österlund, P.; Joensuu, M.; Meunier, F. A.; Butcher, S. J.; Winkler, M. S.; Mollenhauer, B.; Helenius, A.; Gokce, O.; Teesalu, T.; Hepojoki, J.; Vapalahti, O.; Stadelmann, C.; Balistreri, G.; Simons, M., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370 (6518), 856-860.
C, W.; L, W.; Q, Y.; X, W.; J, Z.; X, Y.; Y, Z.; C, F.; D, L.; Y, D.; J, S.; J, G.; X, Y.; Y, W.; X, W.; J, L.; H, Y.; H, L.; Z, Z.; R, W.; P, D.; Y, Z.; F, Y.; W, Z.; N, W.; Y, P.; H, L.; J, F.; C, Q.; W, C.; Q, G.; R, Z.; Y, C.; H, Z., HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nature metabolism 2020, 2 (12).
Gu, Y.; Cao, J.; Zhang, X.; Gao, H.; Wang, Y.; Wang, J.; Zhang, J.; Shen, G.; Jiang, X.; Yang, J.; Zheng, X.; Xu, J.; Zhang, C. C.; Lan, F.; Qu, D.; Zhao, Y.; Xu, G.; Xie, Y.; Luo, M.; Lu, Z., Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv 2020.
Wang, S., Qiu, Z., Hou, Y. et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31, 126–140 (2021)..
L, Z.; CB, J.; H, M.; A, O.; H, P.; BD, Q.; ES, R.; A, P.; A, V.; MS, S.; W, L.; T, I.; C, R.; M, F.; H, C., SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nature communications 2020, 11 (1).
L, Y.; X, W.; KE, P.; C, T.-T.; TP, N.; Y, W.; A, B.; WE, D.; A, D.; C, C.; K, V.; SB, E.; SF, S.; JE, L.; JB, M.; A, R.; A, B.; PC, S.; CA, K.; NV, D.; K, S.; J, L., Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183 (3).
Zhang, W.; Davis, B. D.; Chen, S. S.; Martinez, J. M. S.; Plummer, J. T.; Vail, E., Emergence of a novel SARS-CoV-2 strain in Southern California, USA. Preprint 2021.
Wang, P.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P. D.; Graham, B. S.; Mascola, J. R.; Chang, J. Y.; Yin, M. T.; Sobieszczyk, M.; Kyratsous, C. A.; Shapiro, L.; Sheng, Z.; Nair, M. S.; Huang, Y.; Ho, D. D., Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv 2021.
PANGO lineages. https://cov-lineages.org/global_report.html.
Wu, K.; Werner, A. P.; Moliva, J. I.; Koch, M.; Choi, A.; Stewart-Jones, G. B. E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B. S.; Carfi, A.; Corbett, K. S.; Seder, R. A.; Edwards, D. K., mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv 2021.
Xie, X.; Zou, J.; Fontes-Garfias, C. R.; Xia, H.; Swanson, K. A.; Cutler, M.; Cooper, D.; Menachery, V. D.; Weaver, S.; Dormitzer, P. R.; Shi, P.-Y., Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv 2021.
Novavax, Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. 2021.
Greaney, A. J.; Loes, A. N.; Crawford, K. H. D.; Starr, T. N.; Malone, K. D.; Chu, H. Y.; Bloom, J. D., Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. 2021.
Luan, B.; Wang, H.; Huynh, T., Molecular Mechanism of the N501Y Mutation for Enhanced Binding between SARS-CoV-2’s Spike Protein and Human ACE2 Receptor. bioRxiv 2021.
RL, G.; A, N.; P, C.; J, B.; B, H.; J, M.; G, H.; J, C.; B, M.; V, S.; I, S.; P, K.; AC, A.; J, V. N.; KL, C.; M, D.; G, O.; AE, S.; TR, H.; PJ, E.; RE, H.; NL, K.; J, S.; DR, P.; P, K.; L, S.; DM, S., Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2021.
This web search service is supported by Google Inc.
